Marikki Laiho Lab Research
The sensing, detection and repair of DNA lesions are of vital importance to maintain the genomic integrity and act as barriers against the development of cancer. In advanced cancers, many pathways that govern proper DNA damage control are lost, and conditions prevail which augment the accrual of genetic errors. Several modes of cancer therapies, like radiation and many cytotoxic drugs, exploit the vulnerability of cancer cells incapable of normal damage control. The particular interest of our lab is to understand the regulatory events that prevail in cancer cells, and detect and exploit cancer cell characteristics that could be used as basis of new cancer therapies. The approaches used in the laboratory are aimed to provide novel information on the regulation of cellular DNA damage and tumorigenesis pathways, to identify new targets for therapy, and to apply this knowledge to therapy efforts. The studies aim at a rapid transfer of findings arising from focused mechanistic studies into translational cancer research.
Cellular DNA damage response - p53 and checkpoint responses
Prostate cancer is highly common among men, and often presents with multifocal disease. Of specific relevance to prostate cancers we have shown that the regulation of p53, a key tumor suppressor, and DNA damage checkpoint responses are altered in the human prostate. These findings may indicate that the relaxed damage control could predispose the organ to the highly frequent tumorigenic processes observed clinically. In contrast, primary cancers of the seminal vesicle, an adjacent organ pair under similar hormonal control, are exceedingly rare. By comparing the DNA damage responses of the prostate and the seminal vesicle epithelial cells we have shown their drastically different checkpoint responses to external DNA damage. We have proposed that the prostate might be more prone to the accumulation of genetic aberrations during epithelial regeneration than seminal vesicles due to a weaker ability to enforce the DNA damage checkpoints. These findings highlight the need for better understanding of the molecular mechanisms of prostate tumorigenesis, and may be beneficial in finding out ways to prevent and treat prostate cancer.
RNA Polymerase I and the nucleolus - biology and therapeutic targets
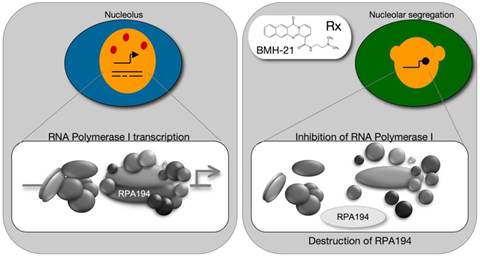
The nucleolus is the site of RNA polymerase I transcription that drives the synthesis of ribosomal (r) RNAs, and processes that engage in processing and assembly of the ribosomes. This is a metabolically highly demanding process, and is on overdrive in cancer cells. We have conducted functional and dynamic proteomic analyses of the nucleolus and quantitative image analysis of the nucleolar responses to proteotoxic, DNA damage and transcription stress. These have provided unique insights into the complexities of nucleolar functions and attest the power of several technological platforms to record and decipher nucleolar processes. We have also recently described a novel small-molecule, BMH-21, that targets Pol I transcription and causes the unprecedented destruction of the Pol I catalytic subunit. The molecule has wide-spectrum anticancer activity in cancer cell lines and in animal tumor models. Further work in the lab will focus on mechanisms of action of BMH-21 on cellular transcriptional programs and efforts towards translating the small-molecule activities to cancer therapies.
|
Recent Publications |
● Peltonen K, Colis L, Liu H, Trivedi R, Moubarek MS, Moore HM, Bai B, Rudek MA, Bieberich CJ, Laiho, M. A targeting modality for destruction of RNA polymerase I that possesses anticancer activity. Cancer Cell 25:77-90, 2014.
● Bai B, Moore HM, Laiho M. CRM1 and its ribosome export adaptor NMD3 localize to the nucleolus and affect ribosomal RNA synthesis. Nucleus, 4:315-325, 2013.
● Bai B, and Laiho, M. Efficient sequential recovery of nucleolar macromolecular components. Proteomics 12:3044-3048, 2012.
● Moore HM, Bai B, Boisvert FM, Latonen L, Rantanen V, Simpson JC, Pepperkok R, Lamond AI, and Laiho M. Quantitative proteomics and dynamic imaging of the nucleolus reveals distinct responses to UV and ionizing radiation. Mol. Cell. Proteomics, 10(10):M111.009241-15, 2011.
● Latonen L, Moore HM, Bai B. Jäämaa S, and Laiho M. Proteasome inhibitors induce nucleolar aggregation of proteasome target proteins and polyadenylated RNA by altering ubiquitin availability. Oncogene 30:790-805, 2011.
● Jäämaa S, Sankila A, Rantanen V, Peltonen K, Järvinen P, af Hällström TM, Ruutu M, Taari K, Andersson LC, Laiho M. Contrasting DNA damage checkpoint responses in epithelium of human seminal vesicle and prostate. Prostate 72:1060-1070, 2012.
● Zhang Z, Yang Z, Jäämaa S, Liu H, Pellakuru LG, Iwata T, af Hällström TM, De Marzo AM, Laiho M. Differential epithelium DNA damage response to ATM and DNA-PK pathway inhibition in human prostate tissue culture. Cell Cycle 10:3545-3553, 2011.
● Jäämaa S, af Hällström TM, Sankila A, Rantanen V, Koistinen H, Stenman U, Zhang Z, Yang Z, De Marzo A, Taari K, Ruutu M, Andersson LC, Laiho M. DNA damage recognition via activated ATM and p53 pathway in non-proliferating human prostate tissue. Cancer Res. 70:8630-8641, 2010.
● Peltonen K, Colis L, Liu H, Jäämaa S, Moore HM, Enbäck J, Laakkonen P, Vaahtokari A, Jones RJ, af Hällström TM, Laiho M. Identification of novel p53 pathway activating small-molecule compounds reveals unexpected similarities with known therapeutic agents. PLoS ONE 5(9): e12996, 2010.
● Kiviharju T, Jäämaa S, Mönkkönen M, Peltonen K, Andersson LC, Medema R, Peehl D, Laiho M. Human prostate epithelium lacks DNA damage-induced checkpoint enforcement by inhibitory Cdk tyrosine 15 phosphorylation. Proc. Natl. Acad. Sci. USA 104:7211-7216, 2007.